© 2020. Paolo Massobrio
The vision
The aim of my research is to study the interplay between the spontaneous activity of neuronal assemblies and the functional topological properties of the network.
Brain is a complex system where thousands of neurons interact by means of an intricate connectivity originating spatio-temporal patterns of electrophysiological activity.
My approach is based on a continuous interaction between computational models and reduced in vitro experimental models based on Micro-Electrode Arrays (MEAs).

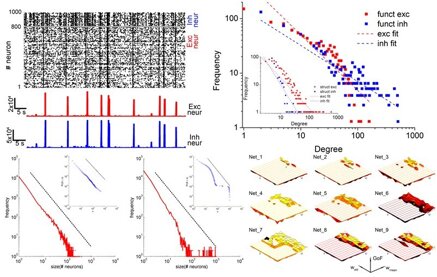
Dynamics vs connectivity
The spontaneous activity of cortical networks is characterized by the emergence of different dynamic states. By combining multi-electrode recordings of spontaneous activity of in vitro cortical assemblies with theoretical models, I am studying how ‘connectivity rules’ drive the network towards different dynamic states, evaluated by means of the Self-Organized Criticality (SOC). Scale-free networks with small-world attributes account better for the variability observed in experimental data, giving rise to different dynamic states.
Interconnected brain regions on-a-chip
Brain is truly three-dimensional (3D) and characterized my interconnected "modules" containing different neuronal types. Such heterogeneity and spatial organization deeply influences the emergent patterns of electrophysiological activity. For these reasons, I am working on the development of innovative in vitro models of 3D interconnected assemblies. Such idea started in 2012, when we co-cultured cortical-thalamic networks over MEAs (doi) and in 2014 when we realized a first 3D hippocampal network over MEAs (doi). Now, I am working on the development of 3D interconnected cortical-hippocampal ensembles as example of brain-on-a-chip where the use of a polymeric confinements in conjunction with a 3D seeding protocol allow the definition of a controllable in vitro model showing realistic dynamics.

Engineered networks

The use of dissociated neuronal cultures allows to drive the connectivity of neuronal networks where topological features can be defined a priori. It is possible to confine neurons in specific regions, defining interconnected assemblies. At the same time, the presence of biocompatibile scaffolds (glass microbeads, hydrogels) allow a 3D growth of the neuronal network. Thanks to the substrate-integrated electrodes of the MEAs, we record the electrophysiological activity and correlate it with the topological properties of the network.
Tools
Since the high complexity of the brain, understanding is not straightforward. For this reason, the use of reduced, controllable in vitro systems mimicking the human brain is away to explore the brain. In addition, such an approach minimizes animal use. The experiments we are performing are based on the use of dissociated neuronal cultures coupled to Micro-Electrode Arrays (MEAs) containing up to 4'000 recording sites.
Indeed, the processing of such an amount of data requires the development of ad hoc suites to extract (quickly and accurately) functional and dynamical properties of the networks.

© 2020. Paolo Massobrio